Understanding Mobile Genetic Elements
At the Dangerfield Lab, we investigate the fundamental mechanisms by which mobile genetic elements—often called “jumping genes”—move and integrate into genomes. These elements make up a large fraction of eukaryotic DNA and play major roles in evolution, genome regulation, and disease.
Focus on R2 Retrotransposons
Our primary focus is the R2 retrotransposon, a mobile element that specifically inserts into the 28S ribosomal RNA (rRNA) gene. After binding its target and using an endonuclease domain to nick the DNA, R2 integrates using a biochemical process known as target-primed reverse transcription (TPRT). In this reaction, the retrotransposon uses the nicked DNA as a primer and its own mRNA transcript as a template for reverse transcription to insert its own genetic material at new copies of 28S rRNA.
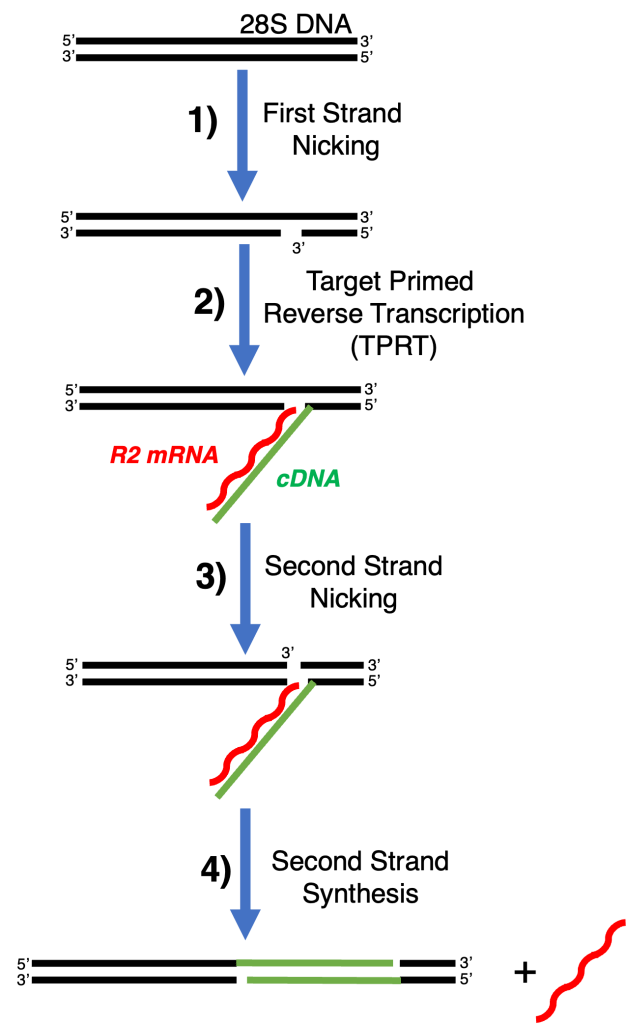
Mechanistic Insights
We use state of the art kinetic methods and next generation DNA sequencing technology to map each step of this process:
- Endonuclease nicking of first strand genomic DNA at a defined rRNA gene site.
- Target-primed cDNA synthesis, where the element’s RNA is copied into DNA by the R2-encoded reverse transcriptase.
- Endonuclease nicking of second strand genomic DNA
- Second-strand synthesis and integration, resulting in a fully inserted retrotransposon.
By dissecting these steps, we aim to understand how enzyme specificity, RNA–DNA interactions, and host factors shape the efficiency and fidelity of retrotransposon integration.
Broader Impact
Our work provides fundamental insights into how genomes evolve, but also explores applications in biotechnology and gene therapy. The precision of R2 integration offers a unique platform for designing tools that harness retrotransposons for programmable gene insertion—enabling new strategies in targeted genome engineering.